Comparing Random Forests and Histogram Gradient Boosting models#
In this example we compare the performance of Random Forest (RF) and Histogram Gradient Boosting (HGBT) models in terms of score and computation time for a regression dataset, though all the concepts here presented apply to classification as well.
The comparison is made by varying the parameters that control the number of trees according to each estimator:
n_estimatorscontrols the number of trees in the forest. It’s a fixed number.max_iteris the maximum number of iterations in a gradient boosting based model. The number of iterations corresponds to the number of trees for regression and binary classification problems. Furthermore, the actual number of trees required by the model depends on the stopping criteria.
HGBT uses gradient boosting to iteratively improve the model’s performance by fitting each tree to the negative gradient of the loss function with respect to the predicted value. RFs, on the other hand, are based on bagging and use a majority vote to predict the outcome.
See the User Guide for more information on ensemble models or see Features in Histogram Gradient Boosting Trees for an example showcasing some other features of HGBT models.
# Author: Arturo Amor <david-arturo.amor-quiroz@inria.fr>
# License: BSD 3 clause
Load dataset#
from sklearn.datasets import fetch_california_housing
X, y = fetch_california_housing(return_X_y=True, as_frame=True)
n_samples, n_features = X.shape
HGBT uses a histogram-based algorithm on binned feature values that can efficiently handle large datasets (tens of thousands of samples or more) with a high number of features (see Why it’s faster). The scikit-learn implementation of RF does not use binning and relies on exact splitting, which can be computationally expensive.
print(f"The dataset consists of {n_samples} samples and {n_features} features")
The dataset consists of 20640 samples and 8 features
Compute score and computation times#
Notice that many parts of the implementation of
HistGradientBoostingClassifier and
HistGradientBoostingRegressor are parallelized by
default.
The implementation of RandomForestRegressor and
RandomForestClassifier can also be run on multiple
cores by using the n_jobs parameter, here set to match the number of
physical cores on the host machine. See Parallelism for more
information.
import joblib
N_CORES = joblib.cpu_count(only_physical_cores=True)
print(f"Number of physical cores: {N_CORES}")
Number of physical cores: 2
Unlike RF, HGBT models offer an early-stopping option (see
Early stopping in Gradient Boosting)
to avoid adding new unnecessary trees. Internally, the algorithm uses an
out-of-sample set to compute the generalization performance of the model at
each addition of a tree. Thus, if the generalization performance is not
improving for more than n_iter_no_change iterations, it stops adding trees.
The other parameters of both models were tuned but the procedure is not shown here to keep the example simple.
import pandas as pd
from sklearn.ensemble import HistGradientBoostingRegressor, RandomForestRegressor
from sklearn.model_selection import GridSearchCV, KFold
models = {
"Random Forest": RandomForestRegressor(
min_samples_leaf=5, random_state=0, n_jobs=N_CORES
),
"Hist Gradient Boosting": HistGradientBoostingRegressor(
max_leaf_nodes=15, random_state=0, early_stopping=False
),
}
param_grids = {
"Random Forest": {"n_estimators": [10, 20, 50, 100]},
"Hist Gradient Boosting": {"max_iter": [10, 20, 50, 100, 300, 500]},
}
cv = KFold(n_splits=4, shuffle=True, random_state=0)
results = []
for name, model in models.items():
grid_search = GridSearchCV(
estimator=model,
param_grid=param_grids[name],
return_train_score=True,
cv=cv,
).fit(X, y)
result = {"model": name, "cv_results": pd.DataFrame(grid_search.cv_results_)}
results.append(result)
Note
Tuning the n_estimators for RF generally results in a waste of computer
power. In practice one just needs to ensure that it is large enough so that
doubling its value does not lead to a significant improvement of the testing
score.
Plot results#
We can use a plotly.express.scatter to visualize the trade-off between elapsed computing time and mean test score. Passing the cursor over a given point displays the corresponding parameters. Error bars correspond to one standard deviation as computed in the different folds of the cross-validation.
import plotly.colors as colors
import plotly.express as px
from plotly.subplots import make_subplots
fig = make_subplots(
rows=1,
cols=2,
shared_yaxes=True,
subplot_titles=["Train time vs score", "Predict time vs score"],
)
model_names = [result["model"] for result in results]
colors_list = colors.qualitative.Plotly * (
len(model_names) // len(colors.qualitative.Plotly) + 1
)
for idx, result in enumerate(results):
cv_results = result["cv_results"].round(3)
model_name = result["model"]
param_name = list(param_grids[model_name].keys())[0]
cv_results[param_name] = cv_results["param_" + param_name]
cv_results["model"] = model_name
scatter_fig = px.scatter(
cv_results,
x="mean_fit_time",
y="mean_test_score",
error_x="std_fit_time",
error_y="std_test_score",
hover_data=param_name,
color="model",
)
line_fig = px.line(
cv_results,
x="mean_fit_time",
y="mean_test_score",
)
scatter_trace = scatter_fig["data"][0]
line_trace = line_fig["data"][0]
scatter_trace.update(marker=dict(color=colors_list[idx]))
line_trace.update(line=dict(color=colors_list[idx]))
fig.add_trace(scatter_trace, row=1, col=1)
fig.add_trace(line_trace, row=1, col=1)
scatter_fig = px.scatter(
cv_results,
x="mean_score_time",
y="mean_test_score",
error_x="std_score_time",
error_y="std_test_score",
hover_data=param_name,
)
line_fig = px.line(
cv_results,
x="mean_score_time",
y="mean_test_score",
)
scatter_trace = scatter_fig["data"][0]
line_trace = line_fig["data"][0]
scatter_trace.update(marker=dict(color=colors_list[idx]))
line_trace.update(line=dict(color=colors_list[idx]))
fig.add_trace(scatter_trace, row=1, col=2)
fig.add_trace(line_trace, row=1, col=2)
fig.update_layout(
xaxis=dict(title="Train time (s) - lower is better"),
yaxis=dict(title="Test R2 score - higher is better"),
xaxis2=dict(title="Predict time (s) - lower is better"),
legend=dict(x=0.72, y=0.05, traceorder="normal", borderwidth=1),
title=dict(x=0.5, text="Speed-score trade-off of tree-based ensembles"),
)
Both HGBT and RF models improve when increasing the number of trees in the ensemble. However, the scores reach a plateau where adding new trees just makes fitting and scoring slower. The RF model reaches such plateau earlier and can never reach the test score of the largest HGBDT model.
Note that the results shown on the above plot can change slightly across runs and even more significantly when running on other machines: try to run this example on your own local machine.
Overall, one should often observe that the Histogram-based gradient boosting models uniformly dominate the Random Forest models in the “test score vs training speed trade-off” (the HGBDT curve should be on the top left of the RF curve, without ever crossing). The “test score vs prediction speed” trade-off can also be more disputed, but it’s most often favorable to HGBDT. It’s always a good idea to check both kinds of model (with hyper-parameter tuning) and compare their performance on your specific problem to determine which model is the best fit but HGBT almost always offers a more favorable speed-accuracy trade-off than RF, either with the default hyper-parameters or including the hyper-parameter tuning cost.
There is one exception to this rule of thumb though: when training a multiclass classification model with a large number of possible classes, HGBDT fits internally one-tree per class at each boosting iteration while the trees used by the RF models are naturally multiclass which should improve the speed accuracy trade-off of the RF models in this case.
Total running time of the script: (0 minutes 55.701 seconds)
Related examples
Sample pipeline for text feature extraction and evaluation
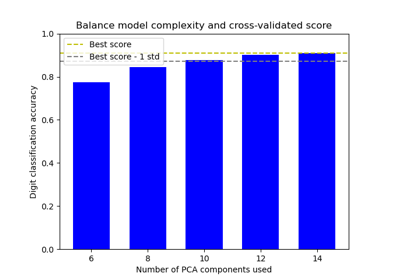
Balance model complexity and cross-validated score